Version 1 (uncompleted)
This version was started to overcome some of the difficulties of dealing with the muModelica AST structure. The flattening procedure was implemented directly in the pyDevs printing code. As the compiler produced pyDevs code for a given coupled DEVS unit, a flattened version would also be produced.
This design was deficient in two regards. First it did not expose any of the DEVS structure to the compiler. This removed the possibility of performing analysis in the compiler. The second deficiency was that the flattening procedure was very simple. A flattened coupled model would simply contain instances of each atomic sub-model in the original model, and instances of flattened models for each coupled sub-model. This is a problem because it makes analysis of the generated code more difficult.
This version was abandoned for a more complex design, but is still partially present in the devsmc.py file.
Version 2
This version intended to expose more of DEVS structure to the compiler. It was also intended to perform more inlining of the flattened structure.
To allow more of the DEVS structure to be exposed to the compiler, new classes were created to capture the structure. This classes are in structure to the Modelica DEVS classes used by muModelica DEVS extension. The class hierarchy can be seen in the following diagram
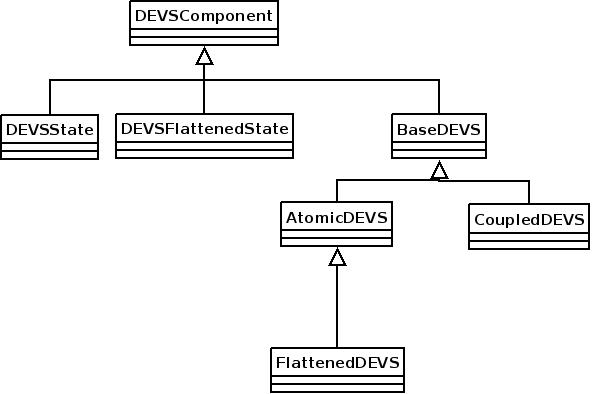
The flattened version of state and DEVS were created to facilitate flattening. As each coupled DEVS is flattened, a DEVSFlattenedState and a FlattenedDEVS instance is created.
Basic design
For each coupled DEVS in the original model, the flattener outputs a flattened version. These flattened versions are not interdependent, though they are dependent on the atomic DEVS that are part of the coupled structure.
The flattening is split into several logical phases.
- Collect models and states from original file and create DEVS objects for them
- Connect the structure of the model in the objects
- Create a topological order of the DEVS objects based on sub-model dependence
-
Flatten the coupled DEVS in reverse topological order
- Create flat instance for the coupled DEVS
- Create flat state instance for the DEVS
- Flatten connections
- add members to the flat state
- add members to the flat model
- create __init__ structure for the flat model
- Print the flattened class
Detailed design
Collecting models
The models are collected from the original modelica file using a simple AST visitor DevsCollector. It traverses the AST and finds class definitions. If the class is a DEVS class, it appends it to either a list of atomics, a list of coupled models, or a list of states.
Connecting the DEVS structure
Once a DEVS object is created to represent the component in the compiler, the AST structure for that component is passed to another visitor(DevsClassCollector) to collect useful information for that component. From this information the models can be linked to their states, and states to their models; and component members can be added to the object. These components include submodels, inputs, outputs, and connections. In addition, connections are stored in a structure of python dictionaries and lists to allow easy access to influencee information.
Topological ordering
Coupled DEVS form a directed acyclic graph of component type dependency with their sub-model structure. From this we can compute a topological ordering. This ordering is necessary when processing a coupled component with coupled sub-components. If the sub components have not been processed, then the higher level component cannot be processed.
Flattening
Each coupled DEVS is flattened in reverse topological order.
First empty objects are created for the flat state and model. Then contents are added to the state and model. The contents of the state consist of records of what atomic components are required for the flat model. This is done by adding all atomic sub-components of the model and recording their name. Then for each coupled sub-component, the components from flat versions of the sub-component are added with updated names. These names are simply the original nameswith the name of the sub-component and _ prepended.
Once members are added, the connections must be flattened. The idea behind the connection flattening is to connect the atomics directly. When a coupled model is involved in the connection, then all atomics involved that coupled model's port internal connections are connected to the other end of the connection being flattened.
With the coupled model flattened, the code gen needs to be able to create an __init__ for the flattened model. This is done by including the __init__ from coupled sub-models with a renaming similar to the renaming done for states. These init methods are recorded and updated to use the new state members.
Printing
Printing is done by the FlattenedDEVS class. First the init method is generated, then the timeAdvance, a method for getting the immediate action, a method getting port connection if an atomic is an influencee of another, the intTransition, and output methods. The extTransition is not currently implemented.
The timeAdvance method will go through each member of the flat model, and checking its timeAdvance (Note: each member of the flat model is an Atomic model at this point). The minimum is found and returned.
The getIStar method is used to find the next component to perform an internal transition. This is done by finding the first component with a ta that matches the timeAdvance of this model.
The isInfluencees method does a lookup in the dictionary structure storing connections for the flat model. If the model is influenced then a list of tuples of in/out ports is returned.
The intTransition method will first execute the intTransition of the immediate component. Then it checks each other component, if its a influencee, an external transition is performed for it, otherwise its elapsed time is incremented.
The output method will go through all the ports of the immediate component that are connected to one of the self ports, and poke the output to the correct port.